Non-genetic differences underlie variability in proliferation among esophageal epithelial clones
Reyes Hueros RA, Gier RA, Shaffer SM
BioRxiv. 2023 June. doi: https://doi.org/10.1101/2023.05.31.543080
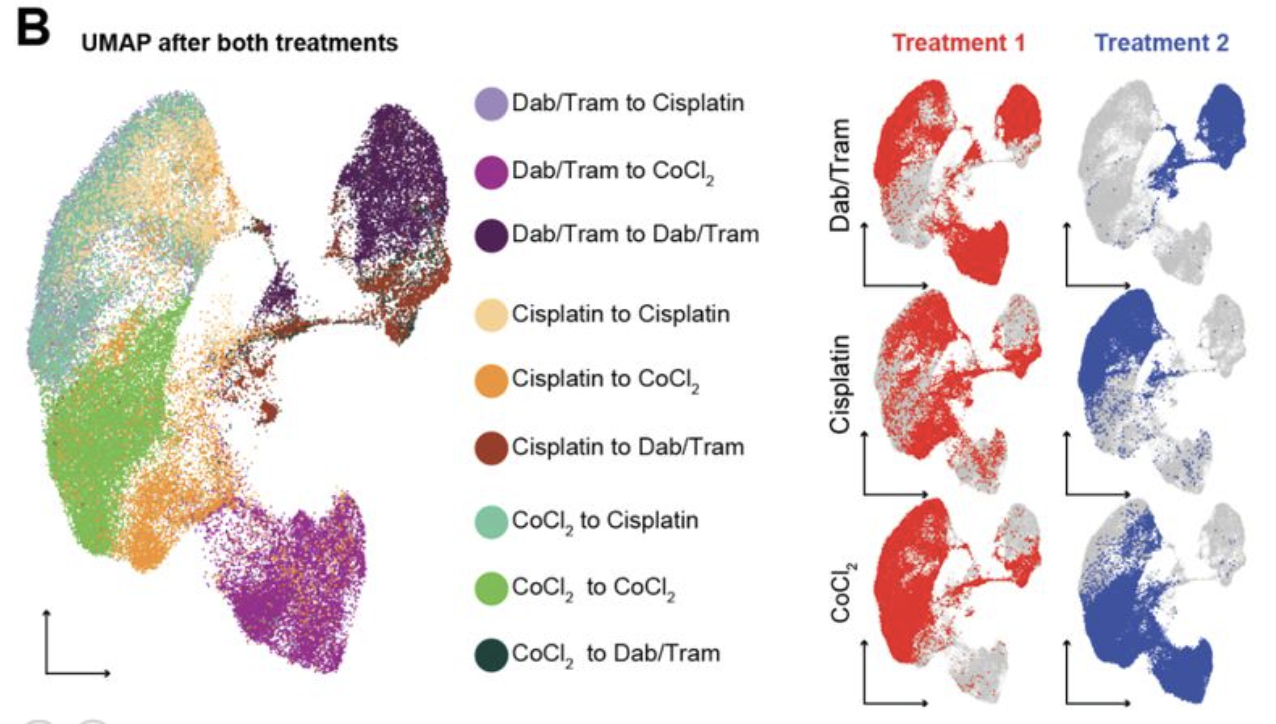
Clonal differences underlie variable responses to sequential and prolonged treatment
Schaff DL*, Fasse AJ*, White PE, Vander Velde R, Shaffer SM *equal contributions
Cell Systems. 2024 March. doi: https://doi.org/10.1016/j.cels.2024.01.011 PDF
Clonal cell states link Barrett’s esophagus and esophageal adenocarcinoma
Gier RA, Reyes Hueros RA, Rong J, DeMarshall M, Karakasheva TA, Muir AB, Falk GW, Zhang NR, Shaffer SM
BioRxiv. 2023 Jan. doi: https://doi.org/10.1101/2023.01.26.525564
Integrating mutational and nonmutational mechanisms of acquired therapy resistance within the Darwinian paradigm
Vander Velde R, Shaffer SM, Marusyk A
Trends in Cancer. 2022 June. doi: https://doi.org/10.1016/j.trecan.2022.02.004
Subcellular Detection of SARS-CoV-2 RNA in Human Tissue Reveals Distinct Localization in Alveolar Type 2 Pneumocytes and Alveolar Macrophages.
Acheampong KK, Schaff DL, Emert BL, Lake J, Reffsin S, Shea EK, Comar CE, Litzky LA, Khurram NA, Linn RL, Feldman M, Weiss SR, Montone KT, Cherry S, Shaffer SM
mBio. 2022 February 8. doi: 10.1128/mbio.03751-21 PDF | pubmed
Memory sequencing reveals heritable single cell gene expression programs associated with distinct cellular behaviors.
Shaffer SM*, Emert B*, Reyes-Hueros RA, Cote C, Harmange G, Schaff G, Sizemore AE, Gupte R, Torre E, Singh A, Basset DS, Raj A *equal contributions
Cell. 2020 August 20. doi: 10.1016/j.cell.2020.07.003 PDF | pubmed
Validation of a next-generation sequencing oncology panel optimized for low input DNA.
Sussman RT*, Shaffer S*, Azzato EM*, DeSloover D, Farooqi MS, Meyer A, Lieberman DB, Bigdeli A, Paolillo C, Ganapathy K, Sukhadia S, Rosenbaum JN, Daber RD, Morrissette JJD. *equal contributions
Cancer Genetics. 2018 December. journal website
SAVER: Gene expression recovery for single-cell RNA sequencing.
Huang M, Wang J, Torre E, Dueck H, Shaffer S, Bonasio R, Murray JI, Raj A, Li M, and Zhang NR.
Nature Methods. 2018 June 25. pubmed
Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance.
Shaffer SM, Dunagin MC, Torborg SR, Torre EA, Emert B, Krepler C, Beqiri M, Sproesser K, Brafford PA, Xiao M, Eggan E, Anastopoulos IN, Vargas-Garcia CA, Singh A, Nathanson KL, Herlyn M, Raj A.